线性权值递减的粒子群优化算法寻找蛋白质最优吸附取向-java
一点点的改进
1. 简介
粒子群优化(Particle Swarm Optimization, PSO),又称粒子群演算法、微粒群算法,是由 J. Kennedy 和 R. C. Eberhart 等[1]于1995年开发的一种演化计算技术,来源于对一个简化社会模型的模拟。其中“群(swarm)”来源于微粒群符合 M. M. Millonas 在开发应用于人工生命(artificial life)的模型时所提出的群体智能的5个基本原则。“粒子(particle)”是一个折衷的选择,因为既需要将群体中的成员描述为没有质量、没有体积的,同时也需要描述它的速度和加速状态。
PSO 算法最初是为了图形化地模拟鸟群优美而不可预测的运动。而通过对动物社会行为的观察,发现在群体中对信息的社会共享提供一个演化的优势,并以此作为开发算法的基础[1]。通过加入近邻的速度匹配、并考虑了多维搜索和根据距离的加速,形成了 PSO 的最初版本。之后引入了惯性权重w来更好的控制开发(exploitation)和探索(exploration),形成了标准版本。为了提高粒群算法的性能和实用性,中山大学、(英国)格拉斯哥大学等又开发了自适应(Adaptive PSO)版本[2]和离散(discrete)版本[3]
PSO 算法属于一种万能启发式演算法,能够在没有得知太多问题资讯的情况下,有效的搜寻具有庞大解空间的问题并找到候选解,但同时不保证其找到的最佳解为真实的最佳解。

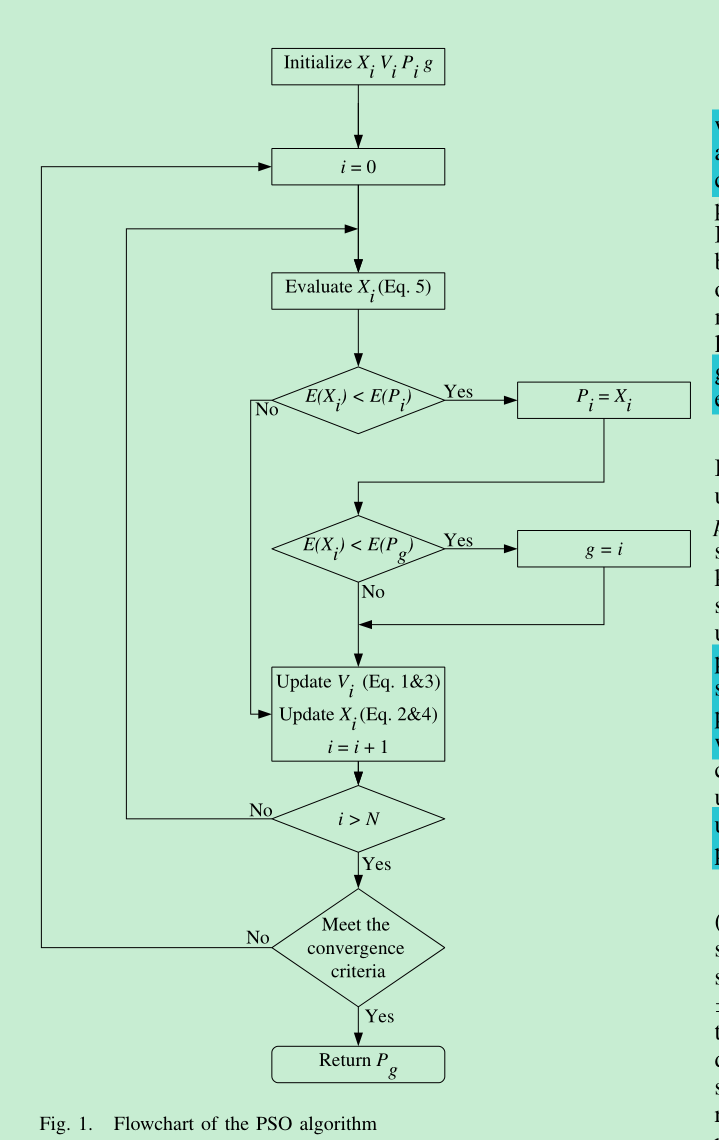
2. 线性权值递减
对于w(惯性权重), 开始时倾向寻找全局最大值, 结束时寻找局部最优值
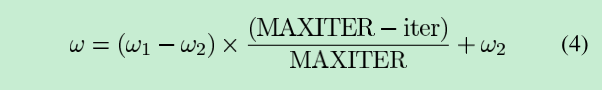
对于c1(自我认知), 开始时权重较大, 结束时权重较小; 值越大越自我
对于c2(社会认知), 开始时权重较小, 结束时权重较大; 值越大越社会
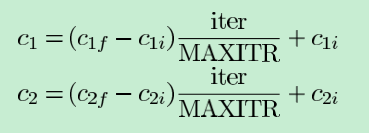
Self-Organizing Hierarchical Particle Swarm Optimizer With Time-Varying Acceleration Coefficients
论文推荐参数:
w: 0.9-0.4;
c1: 2.5-0.5;
c2: 0.5-2.5;
HPSO-TVAC
https://personal.ntu.edu.sg/epnsugan/
3.算法
分为2个部分: Bash脚本调用自定义函数和gromacs建模, java计算部分
每一步保存所有粒子的当前值, step-id.dat
最后保存每一步的最优值, gene.dat
迭代终止条件:运行200次, 或者连续10次最优值相差0.000001; 即认为迭代终止
对于运行em过程, 使用队列控制并行进程数
源码:https://github.com/shxiaj/javaPso
java计算工程文件+bash建模运行脚本
3.1 Bash建模
#!/usr/bin/bash
surfaceFile=sam_cooh.gro
proteinFile=protein.gro
dist2Bottom=0.05
distPro2Surf=0.25
surfTopArgs=("SAM 372" "SBM 28")
surfName="SAM SBM"
postiveCharges=0
negativeCharges=32
X0=$1
X1=$2
X2=$3
X3=$4
X4=$5
wd=$PWD
if [ ! -d "$wd/part/p$6/" ]; then
mkdir -p $wd/part/p$6/
cp -rf $wd/model/* $wd/part/p$6/
else
rm $wd/part/p$6/\#*\#
rm $wd/part/p$6/*/\#*\#
cp -f $wd/model/topol.top $wd/part/p$6/
fi
cd $wd/part/p$6/
########################################################################
# $1 输入文件名; $2 输出文件名; $3 移动后到底边距离(nm)
function g_adjPlus {
trans=`sed '$d' $1 | awk -v dist="$3" 'BEGIN {FIELDWIDTHS="36 8"; min = 100;}
NR > 2 { if ($2 < min) { min = $2; } }
END { trans = dist - min; print trans; }'`
gmx editconf -f $1 -o $2 -translate 0 0 $trans
}
# $1 输入文件名; $2 额外厚度默认为0; return 高度+额外
function g_height {
sed '$d' $1 |
awk -v dist="$2" '
BEGIN {
FIELDWIDTHS="36 8";
min = 100;
max = -100;
}
NR > 2 {
if ($2 < min) {
min = $2;
} else if ($2 > max) {
max = $2;
}
}
END {
thickness = max - min + dist + 0.02;
print thickness;
}'
}
# 获取gro文件的box大小, 输入文件名
function g_boxSize {
awk 'END {printf("%10.5f%10.5f%10.5f",$1,$2,$3)}' $1
}
# $1 组名; $2 gro文件名
function g_findIndex {
echo q | gmx make_ndx -f $2 -o 2>/dev/null |
awk -v resName="$1" '
BEGIN{ FIELDWIDTHS = "3 1 20"; split(resName, s);}
{
for (i in s) {
if ($3 ~ s[i]) { idx[s[i]]=$1;}
}
} END {for (i in idx) {print idx[i]}}'
}
# 获取能量组, 不可调
function getEneGroup {
echo -e "\n\n" | gmx energy -f ./em/em.edr -sum 2>&1 |
awk 'NF == 4 {
for(i=1;i<=NF;i++) {
if ($i ~ "Coul-SR:Protein-Surface") {print $(i-1);}
else if($i ~ "LJ-SR:Protein-Surface") {print $(i-1);}
}
}'
}
# $1 fileName; $2 dist(nm)
function g_boxzPlus {
boxz=`sed '$d' $1 | awk -v dist="$2" 'BEGIN {FIELDWIDTHS="36 8"; max = -50}
NR > 2 {if ($2 > max) max = $2} END {print max+dist}' $1`
sed -i -e "\$s/[0-9]*.[0-9]*\$/${boxz}/" $1
}
function gmxem {
gmx grompp -f em.mdp -c ions.gro -p topol.top -o ./em/em.tpr -po ./em/emout.mdp -n
gmx mdrun -v -deffnm ./em/em -nt 1
}
function g_mer {
if [ -z $2 ]; then p4="ptw4.gro"; else p4=$2; fi
if [ -z $1 ]; then echo "No surface's .gro file"; return; else surf=$1; fi
sed '$d' $p4 > box.gro
sed '1,2d' $surf >> box.gro
line_number=$((`cat box.gro|wc -l` - 3))
sed -i -e "2s/.*/${line_number}/1" box.gro
}
########################################################################
# type gmx5 >/dev/null 2>&1 || { echo >&2 "Gromacs5.0 version not exist! Use System defalut version"; alias gmx5="gmx"; }
# surface preparation
g_adjPlus $surfaceFile surf.gro $dist2Bottom
surfBoxThickness=`g_height surf.gro $dist2Bottom`
surfBoxEdges=($(g_boxSize $surfaceFile))
# protein preparation
gmx editconf -f $proteinFile -o ptw1-1.gro -rotate $X0 $X1 $X2
proBoxThickness=`g_height ptw1-1.gro 1.78`
gmx editconf -f ptw1-1.gro -o ptw1-2.gro -box ${surfBoxEdges[0]} ${surfBoxEdges[1]} $proBoxThickness
gmx editconf -f ptw1-2.gro -o ptw1-3.gro -translate $X3 $X4 0
g_adjPlus ptw1-3.gro ptw2.gro $distPro2Surf
gmx solvate -cp ptw2.gro -cs spc216.gro -o ptw3.gro -p topol.top
# box Setting
gmx editconf -f ptw3.gro -o ptw4.gro -translate 0 0 $surfBoxThickness
g_mer surf.gro ptw4.gro
for (( i = 0; i < ${#surfTopArgs[@]}; i++ )); do
echo -e ${surfTopArgs[$i]} >> topol.top
done
# ions and em
mkdir ion em
gmx grompp -f ion.mdp -c box.gro -p topol.top -o ./ion/ions.tpr -po ./ion/ionout.mdp -maxwarn 1
g_findIndex SOL box.gro | gmx genion -s ./ion/ions.tpr -o ions.gro -p topol.top -np $negativeCharges -pname NA -pq 1 -nn $postiveCharges -nname CL -nq -1
g_boxzPlus ions.gro 0.05
surfId=($(g_findIndex "$surfName" ions.gro))
idxSelect=`for i in ${surfId[@]}; do printf "%d | " $i;done`
idxSelect=${idxSelect::-2}
echo -e "$idxSelect \n q" | gmx make_ndx -f ions.gro -o
lastId=$(($(grep -e "\[.*\]" index.ndx | wc -l) - 1))
echo -e "name $lastId Surface\n q" | gmx make_ndx -f ions.gro -o -n
gmxem
echo -e "`getEneGroup`\n\n" | gmx energy -f ./em/em.edr -sum
awk 'END{print $2}' energy.xvg > ene.dat
3.2 执行脚本
package xyz.shxiaj.pso;
import java.io.File;
import java.io.IOException;
import java.nio.file.Files;
import java.nio.file.Paths;
import java.util.ArrayList;
import java.util.List;
/**
* @Author shxiaj.github.io
* @Date 2022/10/26 13:40
*/
public class ScriptOperation {
public static final String BASH = "bash";
public static final String SCRIPTFILE = "psoem.sh";
public static final String LOGDIR = "./runLog/%d.log";
public static final String ENEDAT = "./part/p%d/ene.dat";
public static Process runEm(double[] variable, int id) throws IOException {
List<String> scriptArgs = new ArrayList<>();
scriptArgs.add(BASH);
scriptArgs.add(SCRIPTFILE);
for (double var : variable) {
scriptArgs.add(String.valueOf(var));
}
scriptArgs.add(String.valueOf(id));
File logFile = new File(String.format(LOGDIR, id));
ProcessBuilder processBuilder = new ProcessBuilder(scriptArgs);
processBuilder.redirectOutput(logFile);
// processBuilder.redirectInput(logFile);
processBuilder.redirectError(logFile);
return processBuilder.start();
}
public static double readDat(int id) throws IOException {
String filePath = String.format(ENEDAT, id);
String datString = Files.readString(Paths.get(filePath));
return Double.parseDouble(datString);
}
}
3.3 pso算法过程
package xyz.shxiaj.pso;
import java.io.FileWriter;
import java.io.IOException;
import java.util.*;
/**
* @Author shxiaj.github.io
* @Date 2022/10/26 10:03
*/
public class Pso {
private double[] gX = new double[Particle.DIMENSION];
private double gFitness;
// setting args for PSO
public static final int CORES = 28;
public static final double PRECISION = 0.000001;
private static final int particleNum = 200;
private static final int N = 200;
private static final double c1i = 2.5;
private static final double c1f = 0.5;
private static final double c2i = 0.5;
private static final double c2f = 2.5;
private static final double wmax = 0.9;
private static final double wmin = 0.4;
private static final String fitDat = "./dat/gene.dat";
private static final String stepDat = "./dat/step-%d.dat";
private final Random random = new Random();
private final List<Particle> parts = new ArrayList<>();
private final List<double[]> allgX = new ArrayList<>();
private final List<Double> allgFitness = new ArrayList<>();
/**
* initial all particle
*/
public void initialParts() {
for (int i = 0; i < particleNum; i++) {
Particle p = new Particle(i);
parts.add(p);
}
}
/**
* update partBest and run em
*/
public void updatePartBest() throws Exception {
Deque<Particle> queue = new ArrayDeque<>();
int i = 0;
while (i < particleNum) {
while (queue.size() < CORES) {
Particle p = parts.get(i);
p.execFitness();
queue.offer(p);
i++;
}
Particle p = queue.poll();
p.process.waitFor();
p.updatePartBest();
}
while (!queue.isEmpty()) {
Particle p = queue.poll();
p.process.waitFor();
p.updatePartBest();
}
}
/**
* update globalBest
*/
public void updateGlobalBest() {
double currBestFitness = Double.MAX_VALUE;
int bestIndex = 0;
// find bestValue and log bestIndex
for (int i = 0; i < particleNum; i++) {
if (parts.get(i).pFitness < currBestFitness) {
currBestFitness = parts.get(i).pFitness;
bestIndex = i;
}
}
// update globalBestValue and globalBest
if (currBestFitness < gFitness) {
gFitness = currBestFitness;
gX = parts.get(bestIndex).pX.clone();
}
allgFitness.add(gFitness);
allgX.add(gX.clone());
}
/**
* update particle Velocity and coord
*/
public void updateVAndX(int n) {
for (Particle p : parts) {
// update velocity for every dimension
for (int i = 0; i < Particle.DIMENSION; i++) {
// Linearly-Decreasing Inertia Weight
double w = wmax - (wmax - wmin) * n / N;
// Time-Varying Acceleration Coefficients
double c1 = c1i + (c1f - c1i) * n / N;
double c2 = c2i + (c2f - c2i) * n / N;
double v = w * p.V[i] + c1 * random.nextDouble() * (p.pX[i] - p.X[i])
+ c2 * random.nextDouble() * (gX[i] - p.X[i]);
p.updatePartVAndV(i, v);
}
}
}
public boolean isConverge() {
int size = allgFitness.size();
if (size >= 10) {
for (int i = size - 2; i > size - 11; i--) {
if (Math.abs(allgFitness.get(i) - allgFitness.get(i + 1)) >= PRECISION) {
return false;
}
}
return true;
} else {
return false;
}
}
public void writerFile() throws IOException {
FileWriter fw = new FileWriter(fitDat, false);
for (int i = 0; i < allgFitness.size(); i++) {
String s = String.format("%5d%15.6f%10.3f%10.3f%10.3f%8.3f%8.3f", i, allgFitness.get(i),
allgX.get(i)[0], allgX.get(i)[1], allgX.get(i)[2], allgX.get(i)[3], allgX.get(i)[4]);
fw.write(s);
fw.write(System.getProperty("line.separator"));
}
fw.flush();
fw.close();
}
public void writerEveryStep(int n) throws IOException {
String datPath = String.format(stepDat, n);
FileWriter fw = new FileWriter(datPath, false);
for (Particle p : parts) {
fw.write(p.toString());
fw.write(System.getProperty("line.separator"));
}
fw.write(String.format("step-%d: gX = %10.3f%10.3f%10.3f%8.3f%8.3f", n, gX[0], gX[1], gX[2], gX[3], gX[4]));
fw.write(System.getProperty("line.separator"));
fw.write(String.valueOf(gFitness));
fw.write(System.getProperty("line.separator"));
fw.flush();
fw.close();
}
public void run() throws Exception {
initialParts();
for (int i = 0; i < N; i++) {
updatePartBest();
updateGlobalBest();
writerEveryStep(i);
updateVAndX(i);
if (isConverge()) break;
}
writerFile();
}
public static void main(String[] args) {
Pso pso = new Pso();
try {
pso.run();
} catch (Exception e) {
e.printStackTrace();
}
}
}
3.4 粒子类
package xyz.shxiaj.pso;
import java.io.IOException;
import java.util.Arrays;
import java.util.Random;
/**
* @Author shxiaj.github.io
* @Date 2022/10/26 10:02
*/
public class Particle {
public int id;
// public double cos;
public double fitness;
public Process process = null;
public static final int DIMENSION = 5;
public final double[] X = new double[DIMENSION];
public final double[] V = new double[DIMENSION];
public final double[][] XLim = {{0, 359.9999}, {0, 359.9999}, {0, 359.9999}, {-0.5, 0.5}, {-0.5, 0.5}};
public final double[][] VLim = {{-36, 36}, {-36, 36}, {-36, 36}, {-0.05, 0.05}, {-0.05, 0.05}};
public double[] pX = new double[DIMENSION];
public double pFitness = Double.MAX_VALUE;
// public double pCos = 0;
public void updatePartVAndV(int i, double v) {
if (v < VLim[i][0]) v = VLim[i][0];
if (v > VLim[i][1]) v = VLim[i][1];
V[i] = v;
double x = X[i] + V[i];
if (x < XLim[i][0]) x = XLim[i][0];
if (x > XLim[i][1]) x = XLim[i][1];
X[i] = x;
}
public void updatePartBest() throws IOException {
double fitness = takeFitness();
if (fitness < pFitness) {
pFitness = fitness;
pX = X.clone();
}
}
public Process execFitness() throws IOException {
this.process = ScriptOperation.runEm(X, this.id);
return this.process;
}
public double takeFitness() throws IOException {
this.fitness = ScriptOperation.readDat(this.id);
return this.fitness;
}
private void initialXAndV() {
Random r = new Random();
for (int i = 0; i < DIMENSION; i++) {
X[i] = r.nextDouble() * (XLim[i][1] - XLim[i][0]) + XLim[i][0];
V[i] = r.nextDouble() * (VLim[i][1] - VLim[i][0]) + VLim[i][0];
}
}
// private void initialV() {
// Random r = new Random();
// for (int i = 0; i < DIMENSION; i++) {
// V[i] = r.nextDouble() * (VLim[i][1] - VLim[i][0]) + VLim[i][0];
// }
// }
public Particle(int id) {
initialXAndV();
// initialV();
this.id = id;
}
@Override
public String toString() {
return "Particle{" +
"id=" + id +
", fitness=" + fitness +
", X=[" + String.format("%10.3f%10.3f%10.3f%8.3f%8.3f", X[0],X[1],X[2],X[3],X[4]) +
"], V=[" + String.format("%10.3f%10.3f%10.3f%8.3f%8.3f", V[0],V[1],V[2],V[3],V[4]) +
"], pX=[" +String.format("%10.3f%10.3f%10.3f%8.3f%8.3f", pX[0],pX[1],pX[2],pX[3],pX[4]) +
"], pFitness=" + pFitness +
'}';
}
}
3.2 格式化最后的数据
function delpunct() {
sed 's/[,:\[]/ /g; s/\]/ /g' $1 | awk '{
printf("%5d",$1);
for (i = 2; i <= NF; i++) {
printf("%10.3f",$i);
}
printf("\n");
}' > $2
}
gmx dipoles -f em.trr -s em.tpr
awk '!/^(#|@)/ {theta = $4 / $5; print $1 " " theta}' Mtot.xvg > dipoles.dat
3.3
javac -d bin ./java/xyz/shxiaj/clpso/*.java
java -cp bin xyz.shxiaj.clpso.Pso
javac -d bin ./java/xyz/shxiaj/clpsoChange/*.java
nohup java -cp bin xyz.shxiaj.clpsoChange.Pso &
java -cp bin xyz.shxiaj.clpsoChange.Pso
3.4
lines=(
1 9 17 25
65 73 81 89
129 137 145 153
193 201 209 217
252 118)